Nutrition and Obesities: Systemic Approaches
UMRS 1269
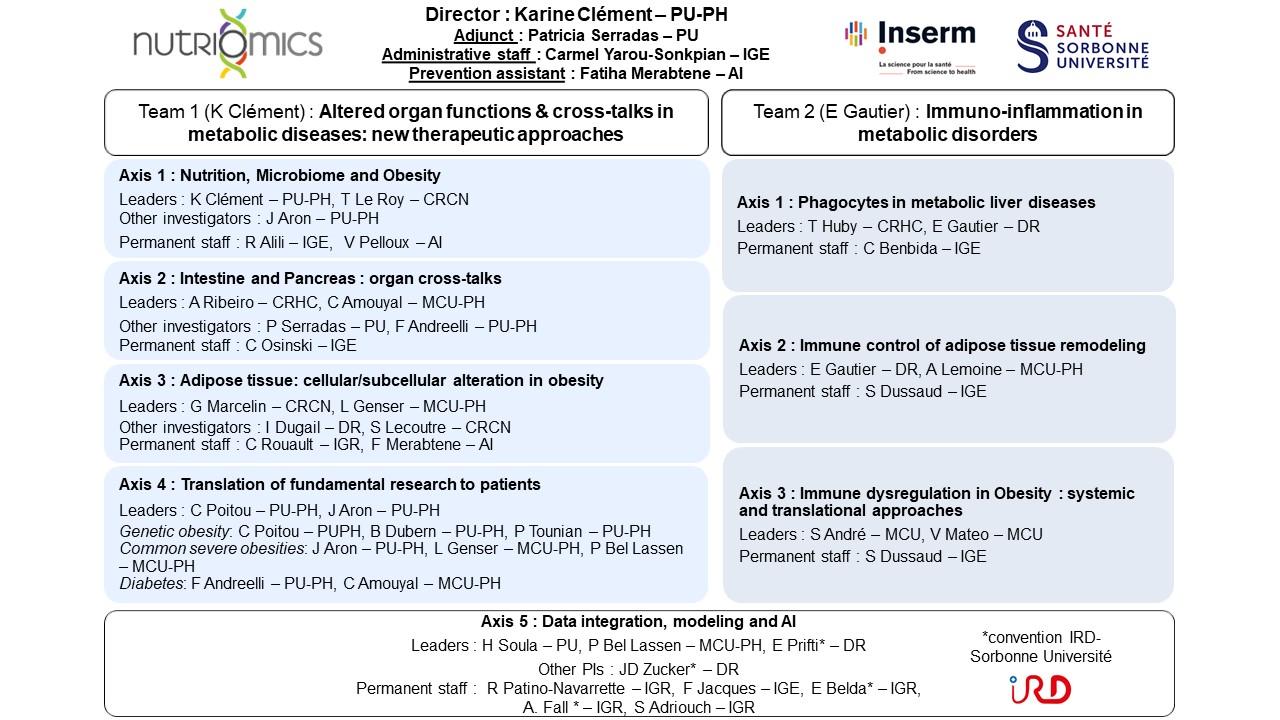
Laboratory headed by Prof. Karine Clément (Director) and Prof. Patricia Serradas (Vice-Director).
For 20 years, our laboratory has been engaged in fundamental and applied research in the field of obesity
and metabolic diseases.
Research Axes
Cross-disciplinary programs spanning nutrition, immunity, adipose biology and data science.
Team 1 : Axis 1 — Nutrition, Microbiome & Metabolic Health
Go to Top ↑
Leaders : K. Clément – PU-PH , T. Le Roy – CRCN
Other investigators : J. Aron – PU-PH
Permanent staff : R. Alili – IGE , V. Pelloux – AI
We investigate how lifestyle and nutrition sculpt the gut microbiota and how microbially derived signals, in turn, shape metabolic and immunoinflammatory health. Building on longitudinal human cohorts assembled over time, we map microbiome features to clinical and immune phenotypes, in close collaboration with Team 2, and launch new large-scale cohorts and nutritional intervention studies to move from association to causation.
Beyond the distal gut, we probe other ecological niches—particularly the jejunum and saliva—to determine their specific contributions to metabolic status. Observational studies relate these communities to clinical readouts, while dedicated strain banks are established from these underexplored sites to enable mechanistic work.
To test causality, we perform human-to-mouse microbiota transfers and targeted experiments in rodents. Guided by species linked positively or negatively with host metabolic health, we evaluate probiotic candidates and assess their capacity to improve metabolic and inflammatory outcomes, paving the way for microbiome-informed nutrition and therapy.
Team 1 : Axis 2 — Intestine & Pancreas : Organ Crosstalks
Go to Top ↑
Leaders : A. Ribeiro – CRHC, C. Amouyal – MCU-PH
Other investigators : P. Serradas – PU, F. Andreelli – PU-PH
Permanent staff : C. Osinski – IGE
This project explores the microbiota–gut–pancreas dialogue to identify new strategies for improving metabolic health. We aim to restore incretin production and enhance beta-cell function by investigating how bacterial metabolites and gut-derived factors influence endocrine signaling in obesity and diabetes.
We will study the impact of microbial metabolites on the sweet taste pathway in human enteroendocrine cells using enteroids derived from subjects with obesity, prediabetes, or type 2 diabetes. These models will allow us to evaluate incretin secretion and gene expression, offering insight into probiotic-based therapeutic approaches.
In parallel, we will identify gut diffusible factors, including miRNAs and peptides, that improve insulin secretion. Using proteomic and transcriptomic analyses, in vitro islet assays, and a microfluidic organ-on-chip system, we will test their relevance after bariatric surgery in both mice and humans.
Finally, co-culture experiments combining enteroids and pancreatic islets on microfluidic chips will clarify the mechanisms of gut–pancreas crosstalk and its role in metabolic regulation.
Team 1 : Axis 3 — Adipose Tissue Remodeling
Go to Top ↑
Leaders : G. Marcelin – CRCN, L. Genser – MCU-PH
Other investigators : I. Dugail – DR, S. Lecoutre – CRCN
Permanent staff : C. Rouault – IGR, F. Merabtene – AI
Adipose tissue flexibility is essential for adapting to energy fluctuations and relies on the coordinated renewal of progenitor, immune, and vascular cells. In obesity, chronic lipid overload disrupts this remodeling, promoting low-grade inflammation and metabolic dysfunction. Our goal is to identify key molecular and cellular alterations driving adipose tissue impairment in severe obesity and to uncover therapeutic strategies to restore metabolic health and prevent weight regain after bariatric surgery.
We will decipher how adipose progenitors contribute to fibrosis and how their fate can be redirected toward healthy adipogenesis. Using cell-specific mouse models, we study PDGFRα+ progenitors and test anti-fibrotic or pro-browning interventions to promote fibrosis resolution during weight loss. Parallel work examines mitochondrial dysfunction in adipocytes, focusing on glutamine metabolism and extracellular vesicle–mediated lipid signaling as drivers of systemic metabolic imbalance.
Leveraging over 1,200 adipose samples from the Nutriomics biobank, we will link molecular phenotypes to clinical outcomes after bariatric surgery, identifying determinants of sustained weight loss. Finally, we explore cellular quality control and extracellular matrix remodeling pathways to restore adipose tissue health and improve long-term metabolic resilience.
Team 1 : Axis 4 — Translation to Patients
Go to Top ↑
Leaders : C. Poitou – PU-PH, J. Aron – PU-PH
Other investigators : B. Dubern – PU-PH, P. Tounian – PU-PH, L. Genser – MCU-PH, P. Bel Lassen – MCU-PH, F. Andreelli – PU-PH, C. Amouyal – MCU-PH
This clinically oriented topic bridges the Nutriomics research group with hospital departments of Nutrition, Diabetology, Surgery, and Gastroenterology at Pitié-Salpêtrière. Its overarching goal is to translate discoveries from fundamental research into clinical applications while building patient cohorts that, in turn, inform mechanistic studies. The focus is on developing innovative diagnostic tools, improving phenotyping, identifying predictive markers, and designing novel therapeutic strategies for obesity and diabetes.
Ongoing studies aim to optimize care for genetic forms of obesity through the ObeRar and Obelisk H2022 programs, assessing long-term outcomes of treatments such as GLP-1 analogs (Wegovy) and setmelanotide. In parallel, clinical trials like DRIFTER and Diabout evaluate metabolic recovery and diabetes remission after bariatric surgery, exploring the contribution of gut microbiota and metformin response.
Finally, advances in diabetology focus on multi-hormonal regulation of insulin secretion and the next generation of incretin-based therapies. Studies on dual and triple GLP-1/GIP/glucagon agonists, intestinal hormone discovery, and nutritional interventions such as the Mediterranean diet aim to improve metabolic outcomes and prevent diabetic cardiomyopathy, reinforcing the translational continuum between bench and bedside.
Team 1 & 2 : Axis 5 — Data Integration, Modeling & AI
Go to Top ↑
Leaders : H. Soula – PU, P. Bel Lassen – MCU-PH, E. Prifti* – DR
Other investigator : J.-D. Zucker* – DR
Permanent staff : R. Patino-Navarrette – IGR, F. Jacques – IGE, E. Belda* – IGR, A. Fall* – IGR
* IRD members
The explosion of biological and clinical data through high-throughput technologies demands interdisciplinary expertise and powerful computational infrastructures. From early microarrays to modern shotgun metagenomics and RNA-seq, each approach generates distinct datasets requiring tailored pipelines and analytical strategies. Since its inception, NutriOmics has remained at the forefront of these advances, pioneering bioinformatics workflows and analytical tools that have enabled novel insights into microbiome and metabolic research.
This topic aims to develop innovative methods in bioinformatics, artificial intelligence, data visualization, and modeling to address key biological and clinical challenges. Closely integrated with all research axes, it supports the analysis of large-scale datasets while driving methodological innovation.
The work is organized around three main axes: computational biology for data-driven exploration of complex biomedical questions; modeling and machine learning for data integration, causality, biomarker discovery, and predictive modeling with interpretable approaches; and finally, pipelines, databases, and high-performance computing to deliver robust, translational tools accessible to both researchers and clinicians.
Team 2 : Axis 1 — Phagocytes in Metabolic Liver Diseases
Go to Top ↑
Leaders : T. Huby – CRHC, E. Gautier – DR
Permanent staff : C. Benbida – IGE
We aim to elucidate how immune cell subsets, particularly liver-resident macrophages (Kupffer cells, KCs), shape the onset and progression of metabolic liver diseases such as steatosis, steatohepatitis, and dyslipidemia. Using transgenic mouse models and leveraging the extensive Nutriomics liver biobank, we will combine mechanistic and translational approaches, including single-nucleus RNA sequencing, to identify key pathways and validate findings in human samples.
Our previous work demonstrated that metabolic alterations during non-alcoholic steatohepatitis (NASH) disrupt KC maintenance and amplify liver injury by sensitizing hepatocytes to cytokine-induced death (Immunity, 2020). Building on this, we will explore how KCs influence hepatocyte demise in acute liver failure and contribute to hepatic fibrosis through interactions with stellate cells. We also investigate how KC dysfunction under hypercholesterolemia perturbs cholesterol metabolism and promotes dyslipidemia.
Finally, in collaboration within a new ANR-funded program, we will investigate neuroimmune regulation of liver inflammation, focusing on how the vagus nerve and parasympathetic signaling modulate KC activity and NASH progression, thus bridging hepatic immunology and neurobiology.
Team 2 : Axis 2 — Immune Control of Adipose Remodeling
Go to Top ↑
Leaders : E. Gautier – DR, A. Lemoine – MCU-PH
Permanent staff : S. Dussaud – IGE
White adipose tissue (WAT) is a highly dynamic organ essential for maintaining metabolic balance. Beyond storing lipids, it can promote energy dissipation through thermogenesis. In obesity, however, WAT undergoes maladaptive remodeling that impairs its flexibility and contributes to metabolic complications such as insulin resistance and type 2 diabetes. Our research focuses on the role of immune cells—particularly macrophage subsets—in regulating WAT remodeling in health and disease.
We will first explore whether macrophages can be harnessed to enhance thermogenic activity, promoting lipid catabolism and reducing fat accumulation. Special attention will be given to the interactions between macrophages and β3-adrenergic signaling pathways that drive thermogenic adipocyte formation in visceral depots, which are closely linked to metabolic dysfunction.
We will also investigate how macrophages influence fibro-inflammatory remodeling of WAT during obesity and after weight loss. By boosting their matrix-degrading capacity, we aim to resolve fibrosis and restore adipose tissue plasticity. Ultimately, this work seeks to uncover macrophage-driven mechanisms that can be targeted to improve WAT function and metabolic health in obesity.
Team 2 : Axis 3 — Immune Dysregulation in Obesities
Go to Top ↑
Leaders : S. André – MCU, V. Mateo – MCU
Permanent staff : S. Dussaud – IGE
The immune system is a complex network of innate and adaptive cells that orchestrate host defense and influence the development of obesity and its related complications, including heightened infection risk and cancer susceptibility. Obesity induces chronic low-grade inflammation, dysbiosis, and insulin resistance, which together disrupt immune homeostasis and contribute to metabolic and inflammatory comorbidities.
Our goal is to understand how obesity-driven alterations in the gut microbiota modulate immune cell function in humans. We will explore dietary interventions designed to restore microbial balance and improve metabolic health, assessing their impact on immune responses through integrative and multi-omic analyses.
Ultimately, this research seeks to clarify the interplay between nutrients, metabolism, and immunity to identify nutritional strategies capable of reestablishing immune system function and resilience in individuals with obesity.